At the start of the pandemic, several studies outlined the effectiveness of radiology imaging for AI-assisted COVID-19 diagnosis through chest X-Ray and mainly Computed Tomography(CT), given the pulmonary involvement in subjects affected by the infection. Even if X-Ray represents a cheaper and most effective solution for large-scale screening, its low resolution led AI models to show lower accuracy than those obtained with CT data.
Several research groups worldwide began to develop deep-learning models for the diagnosis of COVID-19, mainly in the form of deep Convolutional Neural Networks (CNN), applying lung disease analysis from CT scans images. As soon as we started analyzing all the proposed solutions, it was evident that it was impossible to select the most promising ones, due to the use of different and non-comparable architectures, pipelines and datasets. So, we started working on defining a reproducible workflow capable of automating the comparison of state-of-the-art deep learning models to diagnose COVID-19.
The CLAIRE task force on COVID-19
When the pandemic broke out, among the initiatives aimed at improving the knowledge of the virus, containing its diffusion, and limiting its effects, the Confederation of Laboratories for Artificial Intelligence Research in Europe (CLAIRE) task force on AI & COVID-19 supported the set up of a novel European group to study the diagnosis of COVID-19 pneumonia assisted by Artificial Intelligence (AI). The group includes fifteen researchers in complementary disciplines (Radiomics, AI, and HPC), led by Prof. Marco Aldinucci, full professor at the University of Torino Computer Science Dept.
The CLAIRE-COVID19 universal pipeline
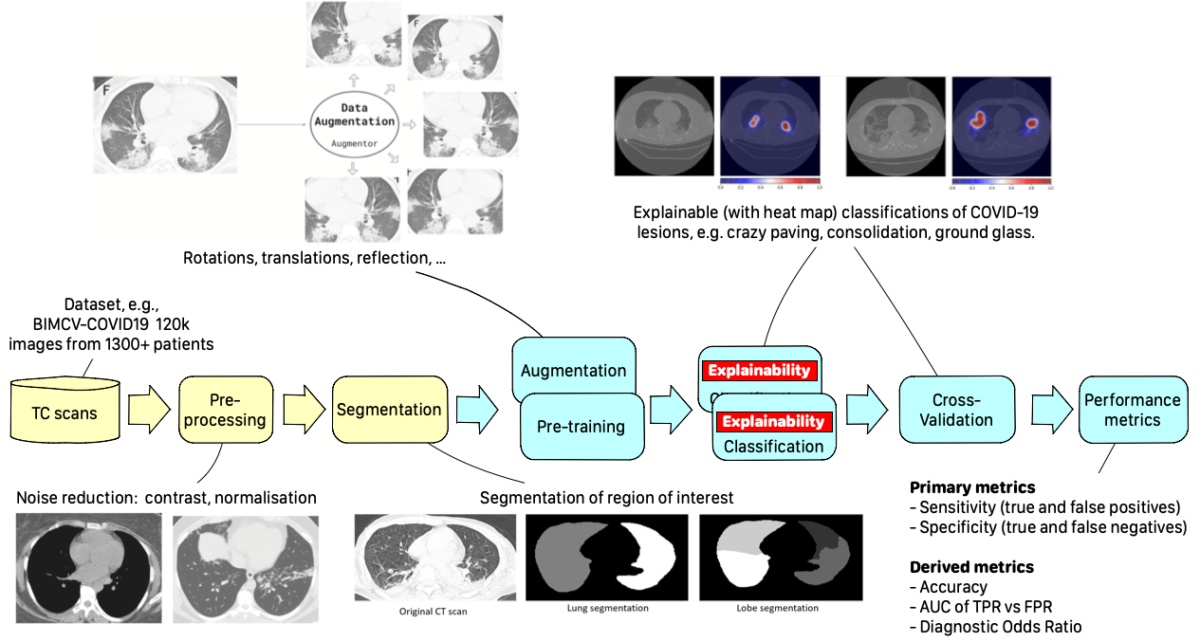
Such collaboration gave birth to the CLAIRE-COVID19 universal pipeline, designed to compare different training algorithms to define a baseline for such techniques and to allow the community to quantitatively measure AI’s progress in the diagnosis of COVID-19 and similar diseases.
The universal pipeline comprises two initial steps: Image Preprocessing and Segmentation. The first applies standard techniques for cleaning and generating variants of training images, while the second uses a DNN-based encoder (e.g., UNet) to isolate a region of interest from the background information (e.g., lungs from other tissues). The final stages are also typical pipeline components implementing Performance metrics and Explainability measures collection.
The core steps are DNN-based. They are Pre-training and Classification. Pre-training aims to generate a first set of weights for the next fine-tuning step, using either an unsupervised technique (e.g., an auto-encoder) or running a supervised training on a different dataset (e.g., ImageNet). The classification step then labels each image with a class identified with a kind of lesion typical of the disease.
Each step can be implemented using different DNNs, generating different variants of the pipeline. We selected the best DNNs that have been experimented in literature for each stage, together with a systematic exploration of the hyper-parameter space, allowing a deeper search for the best model. Moreover, to obtain more consistent results, we applied 5-fold cross-validation to each training process variant.
StreamFlow application
To set up experiments on the pipeline, we chose the most significant dataset publicly available related to COVID-19’s pathology course, i.e., BIMCV-COVID19+, with more than 120k images from 1300 patients. After pre-processing and segmentation phases and a filtering process to remove spurious images, a single training epoch takes on average 5 mins on the resulting dataset.
Running 50 epochs and 5-folds cross-validation on each network configuration translates in a sequential time of about 52 hours for each experiment. A high level of parallelism is needed to run the analysis at scale.
Thanks to StreamFlow and its seamless integration with HPC workload managers, we were able to run the whole spectrum of training configurations for a single DNN (in particular, a DenseNet-121 model) in parallel on the CINECA MARCONI 100 HPC facility. In detail, we explored 12 combinations of hyperparameters with 5-fold cross-validation for a total of 60 experiments. All the experiments ran in parallel on the MARCONI 100 Slurm queue, requesting an NVIDIA Tesla V100 device for each of them.
To further speed up the training, we introduced an early stopping criterion, terminating the training process after ten epochs without improvements in the validation accuracy. With this setting, the whole terminated after ~80 minutes, with a 33.5x speedup compared to a fully sequential run on a single NVIDIA Tesla V100.
I. Colonnelli, B. Cantalupo, R. Esposito, M. Pennisi, C. Spampinato and M. Aldinucci,
“HPC Application Cloudification: The StreamFlow Toolkit,” in 12th Workshop on Parallel Programming and Run-Time Management Techniques for Many-core Architectures and 10th Workshop on Design Tools and Architectures for Multicore Embedded Computing Platforms (PARMA-DITAM 2021), doi: 10.4230/OASIcs.PARMA-DITAM.2021.5.